Irrespective of the aspersions cast by the non-biological alien characters in Terry Bisson’s short story, “They’re Made Out of Meat,” I’m proud to be a carbon-based lifeform. In fact, although I don’t like to boast (I pride myself on my humility and you’ll have to travel a long way to find someone more unpretentious than your humble narrator) I think it’s fair to say that I represent the current peak of evolution for my species (I’m not saying I represent it well, but represent it I do).
The reason I mention this here is that carbon comes in a variety of forms, with 15 isotopes (2 to 16 neutrons), only two of which are stable, along with a mixed bag of allotropes, including amorphous carbon, graphite, graphene, and diamond; buckyballs, carbon nanotubes, carbon nanobuds, and carbon nanofibers; and more esoteric flavors such as ionsdaleite, glassy carbon, linear acetylenic carbon (carbyne), and carbon nanofoam.
In the case of graphene, we’re looking at a two-dimensional sheet of carbon with the atoms arranged in a hexagonal lattice. At the time of this writing, graphene is the strongest material we know. As reported in Nature, graphene transistors, which exhibit a raft of attractive qualities — including tiny size coupled with humongous performance and miniscule power consumption — are considered to be a strong contender for post-silicon electronics.
More importantly in the context of this column is the fact that graphene is a biocompatible material that can be used to implement the latest and greatest in biosensing technology. Recently for example, I enjoyed a video conference with the clever chaps and chapesses at Cardea Bio who are melding graphene, novel chemistry, and electronics to convert biological signals into real-time data.
This all started when I received the following email from the folks at Cardea Bio breathlessly proclaiming what I could only assume to be tremendously exciting news:
Hi Max, we’re excited to share the news of the first global product launch to come out of the Cardea Innovation Partnership Program. Launched in partnership with CRISPR QC, and built upon Cardean Transistor technology, CRISPR-BIND is a rapid and highly sensitive CRISPR quality control tool that can electronically characterize gRNA and CRISPR-Cas interactions as well as other CRISPR QC related assay steps with a precision never seen before.
Accompanying this email was a link to an associated press release: Cardea Bio Announces Global Launch of First Proprietary “Powered by Cardea” Product in Partnership with CRISPR QC. Well, I don’t know about you, but I would have derived just as much understandable information from this email and press release if they had been presented to me in Egyptian hieroglyphics. On the other hand, I have to admit to being intrigued — they had me at “…electronically characterize gRNA and CRISPR-Cas interactions…” — which is what prompted me to request the video conference.
Now, I have to admit that wrapping one’s brain around all this takes a bit of effort, especially if — like yours truly — you are a bear of little brain, but let’s take a run at it and see how well we do.
Let’s start with the fact that the term CRISPR (clustered regularly interspaced short palindromic repeats), pronounced “crisper,” refers to a family of DNA sequences that are found in the genomes of prokaryotic organisms such as bacteria and archaea. These organisms use Cas (CRISPR-associated) proteins to detect and destroy DNA and RNA from attacking bacteriophages (informally referred to as phages), which are viruses that infect bacteria and archaea and replicate within them. The Cas proteins act like molecular scissors, slicing and dicing the invading DNA/RNA at precisely identified points.
Cas9 (CRISPR associated protein 9) is a 160 kilodalton protein that is heavily utilized in today’s genetic engineering applications, including gene editing, which is widely used in basic biological research, the development of biotechnology products, and the treatment of diseases.
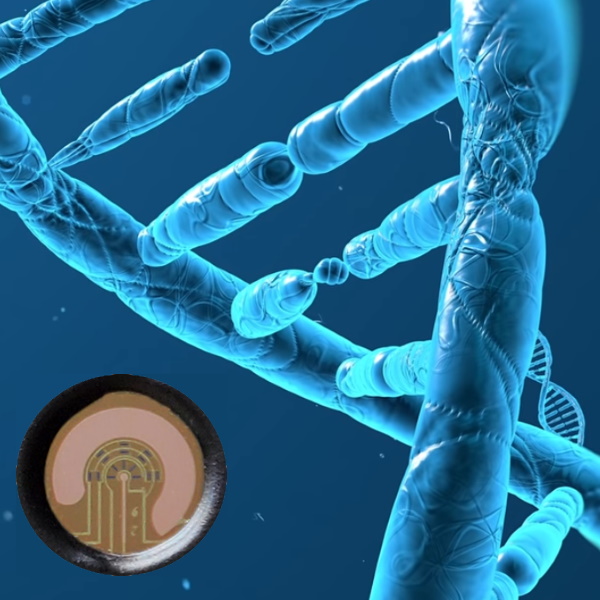
The folks at Cardea Bio have developed biology-gated transistors, a.k.a. Cardean Transistors, that can be used as core modules to build chipsets that can be configured and integrated into thousands of potential products and applications to be used in the field, at the bedside, or on the laboratory bench. One example deployment of this technology is the CRISPR-Chip, which combines thousands of CRISPR-Cas9 molecules with what may be thought of as a graphene field-effect transistor. These CRISPR molecules have been designed to scan through DNA to find specific genes or mutations. Meanwhile, the graphene transistor is super-sensitive to electrically charged materials, like DNA.
When DNA is presented to the CRISPR-Chip, the CRISPR molecules scan it looking for their designated sequence. If they fail to find anything, they will release the DNA. If they do find what they are looking for, they will bind to the DNA, where this binding creates an additional charge on top of the graphene surface. This charge can be sensed, thereby informing the users of the CRISPR-Chip that they’ve found what they were looking for in a matter of minutes.
The folks at Cardea Bio like to talk about their Tech+Bio infrastructure (if the truth be told, it’s hard to get them to stop talking about their Tech+Bio infrastructure). This starts with their Cardean transistors, which — as we’ve already discussed — are composed of biocompatible graphene combined with small bits of biology to create chipsets that can be integrated into thousands of different applications. They’ve also developed versatile hardware that ranges from handheld portable devices that can be run in the field to automated high-throughput systems that live on the lab bench.
But wait, there’s more, because they’ve also developed associated artificial intelligence (AI) and machine learning (ML) software, which allows direct multi-omics data-streams of biological signals to be used to train ML/AI algorithms on how the biology-based sub-systems function. Using a cloud-based system, the guys and gals at Cardea Bio and their partners can access and act on streams of such signals in real-time anywhere in the world.
I can think of all sorts of applications for this sort of technology, from medical monitoring and clinical testing, to breath, sweat, and skin sensors, to food safety, agriculture, and defense, to goodness only knows what. The main problem for the folks at Cardea Bio is that there are simply too many potential projects for them to progress on their own. Thus, they’ve established something called The Cardea Innovation Partnership Program with the goal of providing a diverse set of partners with access to the company’s biology gated transistors and software. The first global product launch to come out of this program is CRISPR-BIND, which provides automated CRISPR Quality Control testing at the comfort of a lab bench. This was developed as a partnership between CRISPR QC and Cardea Bio. Ah ha! Now the aforementioned email and press release start to make sense!
I tell you; I’m constantly amazed at all of the cool technologies that are currently popping up like (genetically modified) mushrooms. And we’re still at the very early days of these technologies. I wonder where this will take us in the coming years (now I’m thinking of Blade Runner and Steel Beach). How about you? Do you have any thoughts you’d care to share?